2023 Summer Research at the NIH
Investigating foundational language models to capture better patient representation
Context
This summer I had the privilege of working on natural language processing and deep learning models in Dr. Zhiyong Lu’s lab under the mentorship of Dr. Qingyu Chen. I was exposed mainly to the HuggingFace libraries as well as Pytorch and Keras architectures. In addition, I learned more about high performance computing cluster and parallelization with GPU’s via NIH’s Biowulf.
The problem statement this summer was to investigate whether foundational language models can become a viable proposed solution to generally capture patient representation in data. This is important because in patient data there are many language ambiguities and inconsistencies such as redundancy and abbreviations of acronyms. The first foray into investigating whether a language model can perform better on generalizing patient representation is by choosing a model and the dataset on which to train on. The model we chose to start this project off is called BERT.
Project
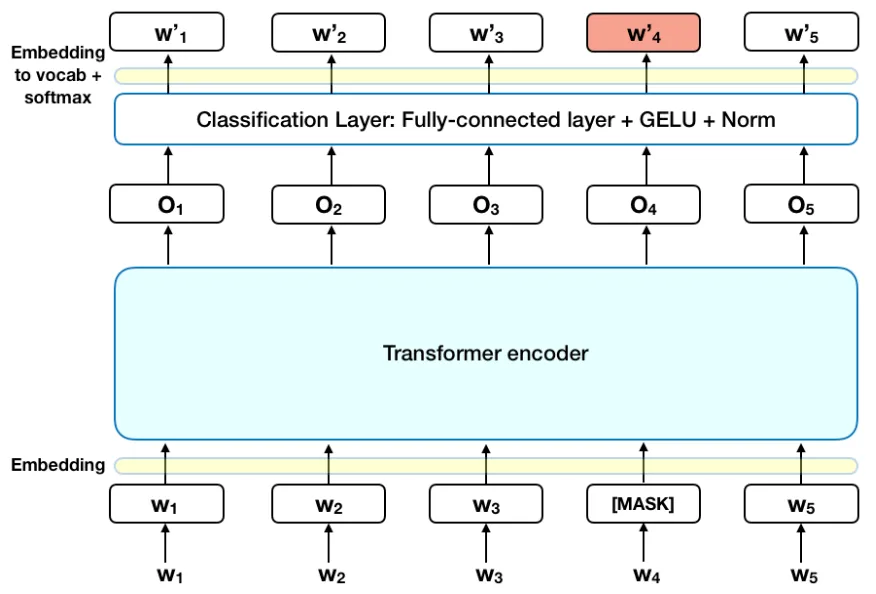
BERT is short for Bidirectional Encoder Representations from Transformers. It has had a lot of success in the biomedical domain with versions such as ClinicalBERT, BioBERT, and PubMedBERT. Although these already exist, we wanted to investigate the viability of using PMC-Patients, a dataset with 167,000 patient summaries extracted from biomedical literature/articles in a data source called PubMed Central. This dataset was found to be more well-rounded compared to its counterpart data sources MIMIC and TREC, which have more elderly populations and patient summary lengths that are either too small or too large. PMC-Patients, with each summary averaging 410 words in length, seemed to be a great starting point for this exploratory analysis.
A bunch of preprocessing and package installation errors in Biowulf later, we began to pretrain a base BERT model (learn from scratch) using the PMC-Patients dataset using MLM.
What are Pretraining Objectives? - MLM and NSP
Masked Language Modeling is an objective where we hide a certain ratio of the token representation of each sentence so the model has to “fill in the blank”. This is supposed to assist the model in giving it more human-like intuition.
Next Sentence Prediction is another pretraining objective that sees how well a model can determine whether or not one sentence comes after another.
Due to time constraints and the exponentially longer training, only MLM was done for this summer. However, the script for MLM and NSP in the pipeline was also developed.
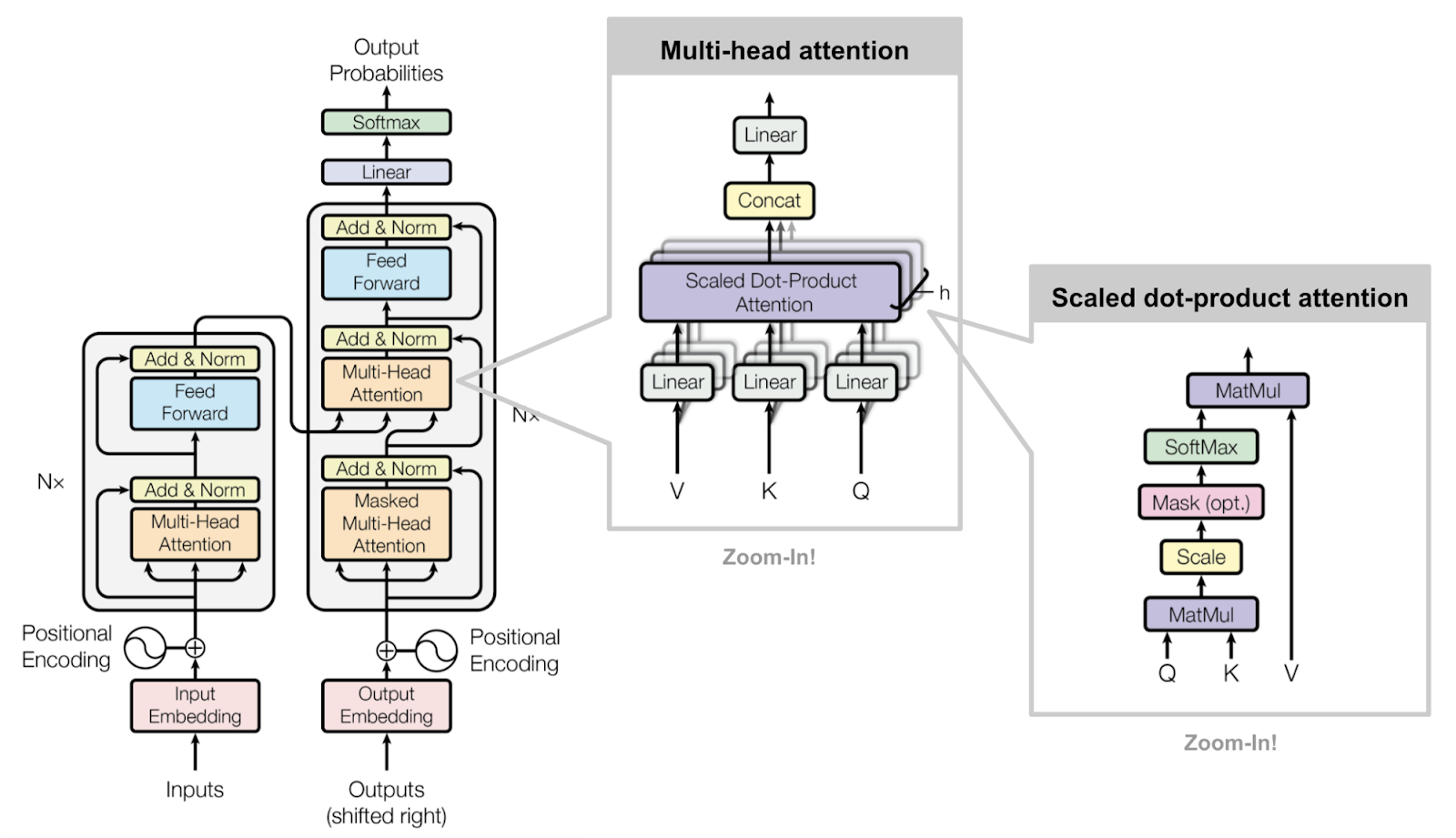
Due to time constraints of my summer internship (10 weeks), I was asked to further evaluate an intermediate model (590/1000 epochs) using 2-3 different downstream tasks compared to its base counterpart trained on a general English corpus for now. These tasks included semantic similarity, named entity recognition, and relation extraction from various datasets. I won’t go too deep into detail of the results, but will explain what each of these downstream evaluation tasks are.
Downstream Tasks and Evaluation
What are Downstream Tasks?
Downstream tasks are evaluation tasks on datasets that have never been seen or learned by the model before. Often, this is used to evaluate how well a model can perform and generalize the knowledge from pretraining, and to show how versatile a model can be when performing tasks. There are two types of downstream tasks: intrinsic and extrinsic.
Intrinsic tasks are tasks done during the pretraining process for internal evaluation of a mdoel for hyperparameter adjustment. Often, this is done using bootstrapping or other statistical methods on the dataset it was trained on.
Extrinsic tasks are tasks done for a certain purpose for evaluation. This is normally done on external datasets the model has never seen before.
Datasets
Two datasets were investigated for similarity and NER/RE respectively: ClinicalSTS and N2C2 ADE. ClinicalSTS is a 2019 dataset from a bioinformatics challenge that consists of sentence pairs rated on a scale of 0-5, 0 meaning no semantic overlap in similarity and 5 meaning complete semantic overlap.
N2C2’s ADE dataset is 505 files from MIMIC that are clinical notes related to Adverse Drug Events and their entities and relationships. The goal of the NER and RE tasks on this dataset is to see how well the model can catch these entities (that can be 1 or more words) and their relationships. The data was split into text files of the clinical narratives and their respective annotations (in .ann files), or dictionaries that has the entities positions in the text. Some preprocessing needed to be done to get csv files to feed in for each task respectively.
Semantic Similarity
The semantic similarity task seeks to evaluate how well a model can detect whether given a sentence pair is similar in meaning or not. This sounds like a simple task, but meaning can be hard to contextualize in a certain domain due to different wording and contexts in different sentences. Ultimately, this is a classification problem, that has two classes that may have very little differences in the interpretation made by the model. In this case for the summer, the pretrained BERT did marginally better on this task but still had a pretty hard time telling the difference between similar and different as classes.
Named Entity Recognition (NER)
The model was able to name more entities than its counterpart which labeled more O’s or Other Tokens rather than entities. However, this can be hard to compare since the base model seemed to have capture a smaller number of entities in general. This may be because they have different tokenizers, BERT baseline has a tokenizer trained on general English while the pretrained BERT has a tokenizer based on PMC-Patients. Further investigation of my preprocessing methods needs to be performed.
Relation Extraction (RE)
The model did not perform as well on relationships even when isolating text with only one relationship or including the whole dataset. Further investigation on my preprocessing method is probably needed to verify the accuracy of my extraction script as well as evaluation of the model performance.
Conclusion of the Summer Project and Future Work
Overall, I had a great time learning about the transformers architecture, BERT, and deep learning from my lab and mentor. It was a challenging project for someone who has never taken a Deep Learning class before, and I learned way more about what goes into training and finetuning a model than I ever would have at my workplace. High performance computing is an amazing privilege as data center strength GPU’s are hard to acquire and expensive to use outside of the NIH.
In the future, this work should be expanded to further hyperparameter tune the model and evaluate the full iteration of the model compared to its biomedical NLP counterparts on the same/other downstream tasks.
Reflection and Other Fun Things
This summer was a great learning experience and I had a lot of fun! There were many summer interns in my cohort for my NIH summer program of all different interests/backgrounds (in addition to interns in my building in different labs). I had a steady lunch group weekly on Fridays where I would explore restaurants in downtown Bethesda, a few lunches with my lab, and even got to take some classes on being resilient facing stress and conflict. All this free NIH content was great for personal development as well as career. Much like a college campus, NIH offers a bunch of services to its interns over the summer that anyone should take advantage of.
We had presentations in our lab for progress updates 2-3 times this summer to practice our presentations and keep Dr. Lu up to date. Despite doing countless presentations in industry, I was fairly nervous because I had never done a research related one before. I learned to adapt quickly to the format and was able to pull my act together before the final one, which I personally feel proud of. I also have a lot to improve on that, as time goes!
I also attended a journal club that had 10-12 other interns for 4 weekly sessions where we would discuss a new paper in a series of papers about the “history” of NLP. Each groups of 3 summer interns had to read and present a paper about NLP and discuss critiques and what the paper was about to their peers. I found this journal club format to be amazing for engagement, as opposed to a lecture style. It helped me learn how to read papers more effectively as well as cross reference their citations, as someone who has never read academic literature.
I would highly recommend anyone interested in Data Science to apply to the GDSSP program at the NIH or SIP for any biomedical summer research.
Related Media
You can find my summer poster, presentation slides, and journal club media here.
References
- Zhao, Z., Jin Q., Chen F., Peng T., Yu S. (2023). PMC-Patients: A Large-scale Dataset of Patient Summaries and Relations for Benchmarking Retrieval-based Clinical Decision Support Systems. ArXiv, abs/2202.13876.
- Chen, Q., Du, J., Hu, Y., Keloth V., Peng, X., Raja, K., Zhang, R., Lu, Z., Xu H. (2023). Large language models in biomedical natural language processing: benchmarks, baselines, and recommendations. ArXiv, abs/2305.16326.
- Henry, S., Buchan, K., Filannino, M., Stubbs, A., & Uzuner, O. (2020). 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. Journal of the American Medical Informatics Association : JAMIA, 27(1), 3–12. https://doi.org/10.1093/jamia/ocz166
- Wang, Y., Fu, S., Shen, F., Henry, S., Uzuner, O., & Liu, H. (2020). The 2019 n2c2/OHNLP Track on Clinical Semantic Textual Similarity: Overview. JMIR medical informatics, 8(11), e23375. https://doi.org/10.2196/23375
- Devlin, J., Chang, M., Lee, K., & Toutanova, K. (2019). BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. ArXiv, abs/1810.04805.